By Cristina-Iulia Bucur, Vrije Universiteit Amsterdam, NL
Scientific publishing methods are often stuck in the past, adhering to formats that are optimized for print use, such as PDF files, rather than digital formats that are more transparent and machine-readable. We need to make use of technological advances around the semantic web and linked data as we redefine the whole publishing process – no longer focusing on the final article but using a fine-grained and networked approach so snippets of information can be accurately tracked together. Read on to discover more!

We currently disseminate, share, and evaluate scientific findings following paradigms of publishing where the only difference from the methods of more than 300 years ago is the medium in which we publish – we have moved from printed articles to digital format, but still use almost the same natural language narrative with long coarse-grained text with complicated structures. These are optimized for human readers and not for automated means of organization and access. Additionally, peer reviewing is the main method of quality assessment, but these peer reviews are rarely published and have their own complicated structure, with no accessible links to the respective articles. With the increasing number of articles being published, it is difficult for researchers to stay up to date in their specific fields, unless we find a way to involve machines as well in this whole process. So, how can we make use of the current technologies to change these old paradigms of publishing and make the process more transparent and machine-interpretable?
In order to address these problems and to better align scientific publishing with the principles of the Web and linked data, my research proposes an approach to use nanopublications – in the form of a fine-grained unifying model – to represent in a semantic way the elements of publications, their assessments, as well as the involved processes, actors, and provenance in general. This research is a result of the collaboration between Vrije Universiteit Amsterdam, IOS Press, and the Netherlands Institute for Sound and Vision for the Linkflows project. The purpose of this project is to make scientific contributions on the Web, e.g. articles, reviews, blog posts, multimedia objects, datasets, individual data entries, annotations, discussions, etc., better valorized and efficiently assessed in a way that allows for their automated interlinking, quality evaluation, and inclusion in scientific workflows. My involvement in the Linkflows project is in collaboration with Tobias Kuhn, Davide Ceolin, Jacco van Ossenbruggen, Stephanie Delbecque, Maarten Fröhlich, and Johan Oomen.
Semantic publishing
One concept that first comes to mind and a proposed solution to make scientific publishing machine-interpretable is “semantic publishing.” This is not a new concept, as its roots are tightly coupled to the notion of the Web, with Tim Berners-Lee mentioning that the semantic web "will likely profoundly change the very nature of how scientific knowledge is produced and shared, in ways that we can now barely imagine" [1]. Despite the fact that semantic publishing is not directly linked to the Web, its progress was highly influenced by the rise of the “semantic web.” As such, in the beginning, it referred to mainly publishing information on the Web in the form of documents that additionally contain structured annotations, so extra information that is parsable by machines in the form of semantic markup (with markup languages like RDFa, for example). This allowed published information on the Web to be machine-interpretable, to the limited extent to which the markup languages allowed. A next step was to use semantic web languages like RDF and OWL to publish information in the form of data objects, together with a specific detailed representation called ontology that is able to represent the domain of the data in a formal (thus machine-interpretable) way. The information published in such structured ways provides not only a “semantic” context through the metadata that describes the information, but also a way for machines to understand the structure and even the meaning of the published information [2,3].
In this way, semantic publishing would allow for the “automated discovery, enables its linking to semantically related articles, provides access to data within the article in actionable form, or facilitates integration of data between papers” [3]. However, despite all the advancements in the semantic web technologies in the past years, semantic publishing is not “genuine” [4] in the sense that the current scientific publishing paradigm has not changed much as we are still using long articles written in natural language that do not contain formal semantics from the start that machines can process and interpret in an automated manner. So, with scientific publishing often stuck to formats optimized for print such as PDF, we are not using the advances that are available to us with technologies around the semantic web and linked data.

Transforming scholarly articles into small, interlinked snippets of data
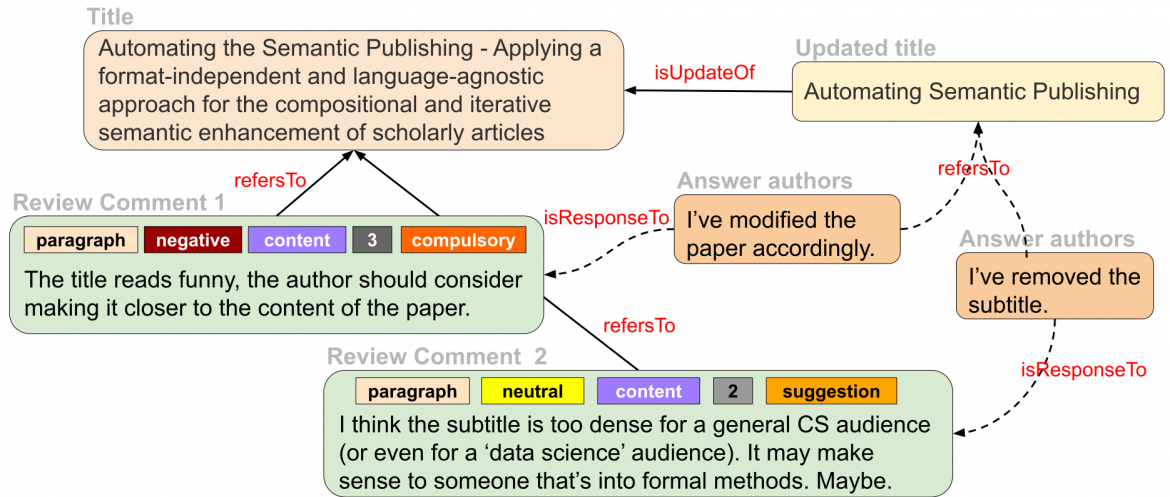
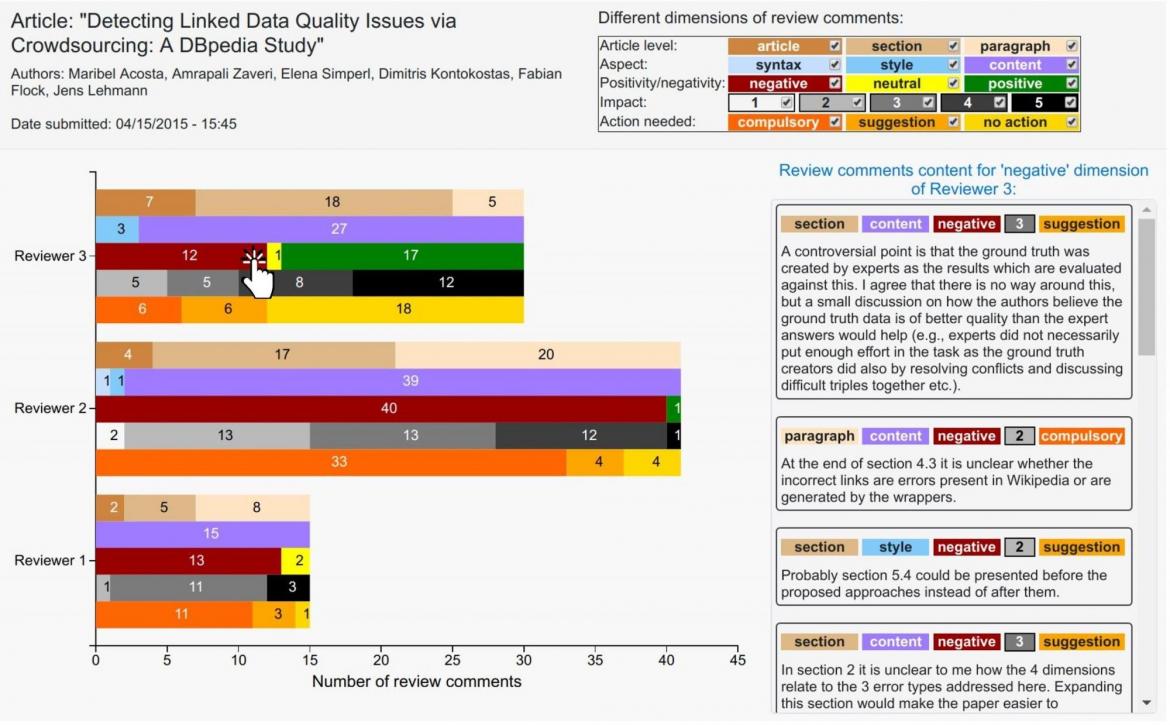
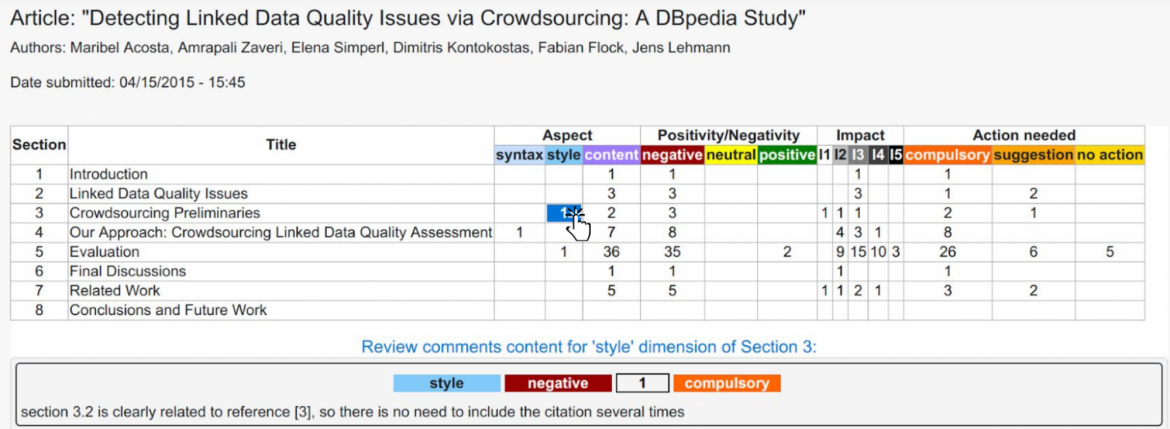
In our approach, we argue for a new system of scientific publishing that contains smaller, fine-grained, formal – machine-interpretable – representations of scientific knowledge that are linked together in a big knowledge network in a web-like fashion in a way that these publications do not need to be necessarily linked to a journal or a traditional publication and can be publication entities by themselves [5,6]. Moreover, semantic technologies make possible the decomposition of traditional science articles into constituent machine-readable parts that are linked not only with one another, but also to other related fine-grained parts of knowledge on the Web following the linked data principles. So, we designed a model for a more granular and semantic publishing paradigm, which can be used for scientific articles as well as reviews.
Figure 1: The scientific publishing process at a granular level: